
Photo: Bryan Goodchild and Rob Carlin
An international team of researchers co-led by Job Dekker, PhD, at UMass Chan Medical School, have identified rules that tell cells how to fold DNA into the tightly packed, iconic X-shaped chromosomes formed during mitosis that help ensure the accurate passing of genetic information between cells during cell division.
Published in the journal Science, these findings illuminate basic biological functions underlying mitosis on a micrometer scale. Understanding how the cells accomplish this critical task may provide important new insights into inheritance and DNA stability and repair, as well as genetic mutations that lead to human diseases such as cancer.
“While we and many others have contributed to the general understanding of how the genome is folded inside chromosomes during mitosis—how cells dynamically acquire their intricate folding arrangement—squeezing several centimeters of genetic information into pairs of complexly arranged sister DNA molecules only a few micrometers in size has been more challenging to unpack,” said Dr. Dekker, Howard Hughes Medical Investigator, the Joseph J. Byrne Chair in Biomedical Research and professor of systems biology. “The work by our team outlines how cells follow a few simple rules to build chromosomes and fold DNA into these tiny, intricate structures during the mitotic phase of cell division.”
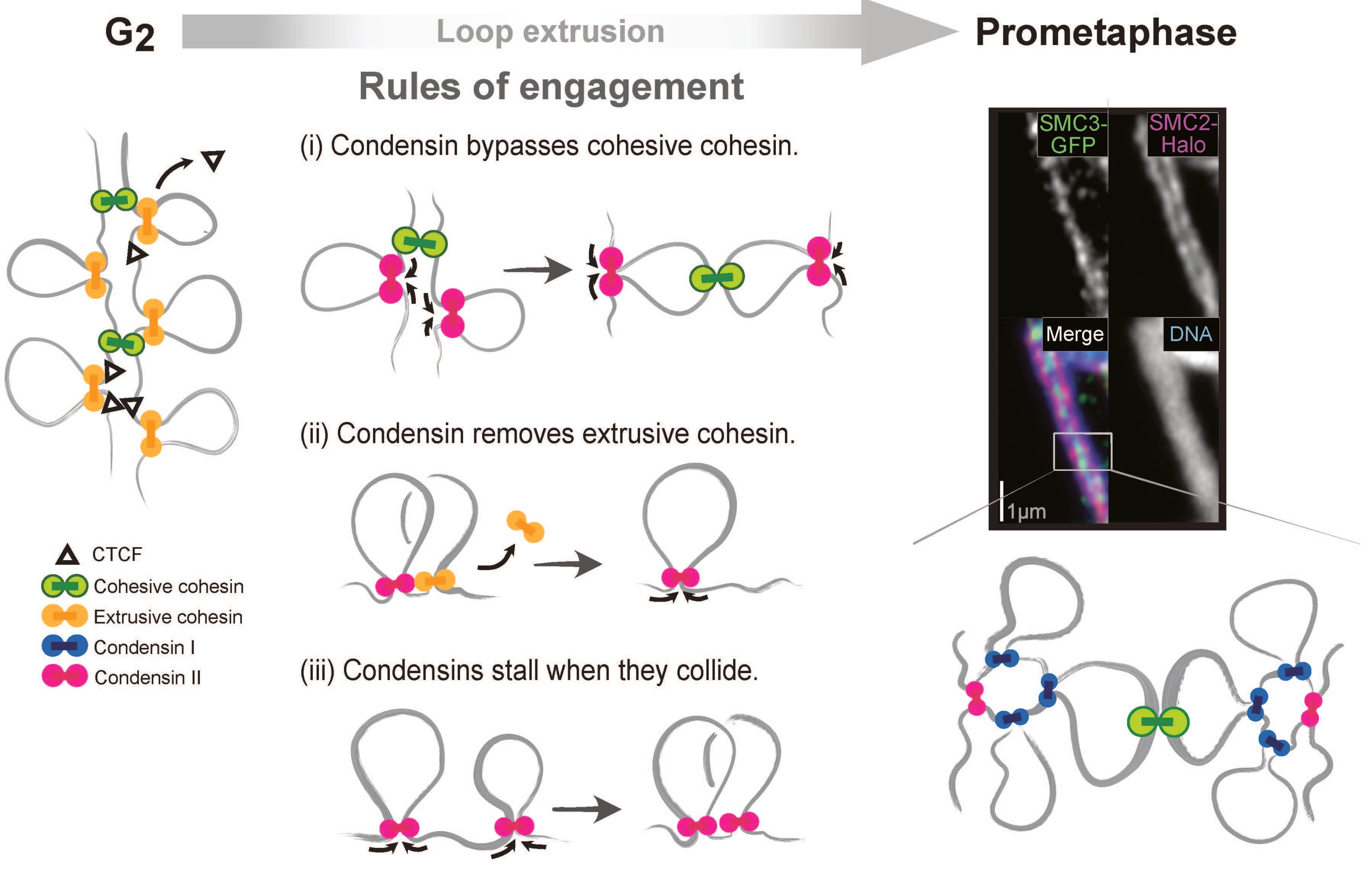
There are three rules of engagement, identifying by Job Dekker, PhD, and Johan Gibcus, PhD, at UMass Chan Medical School, defining how chromosome folding machines interact when they encounter each other along the genome during mitosis: Displacement, Bypassing and Stalling. This video shows what it looks like when the four machines—cohesive cohensin (green circle), extrusive cohensin (yellow circle), condensin I, (blue circle) and condensin II (red circle)— run into each other on the genome during mitsosis.
Video by Johan Gibcus, PhD, and Job Dekker, PhD
Co-leading the study with Dekker are William C. Earnshaw, PhD, professor of chromosome dynamics at the University of Edinburgh; Leonid A. Mirny, PhD, professor in medicine and biomedical physics and physics at the Massachusetts Institute of Technology; and Anton Goloborodko, PhD, group leader at the Institute of Molecular Biotechnology in Vienna.
When we think of chromosomes, many of us imagine the familiar X-shaped structures from high school biology that can be observed in dividing cells during mitosis. These structures are among the most iconic in cellular biology. More importantly, their precise formation is essential for cell division and passing DNA from one cell generation to the next.
Inside each of these tiny, X-shaped chromosomes are small pairs of sister DNA molecules called chromatids, identical halves of the chromosome created when DNA is copied in preparation for cell division during mitosis. The chromatids, joined at the centromere, are several centimeters long, but compacted by the cell to a length of only a few microns. (After cell division there are 92 sister chromatids per cell.) However, how cells fold these tiny strands of DNA into densely compacted, intricate structures has been a mystery to cell biologists and geneticists for more than 150 years.
Graphic: Johan Gibcus, PhD
Work by Dekker and others over the past 40 years has shown that inside each of these chromatids, the DNA is folded as a series of loops and the two sister chromatids are connected to each other all along their length. These loops of DNA are generated in the cell by a set of molecular machines called cohesins and condensins, which are responsible for binding DNA and then pumping out loops of chromatin at a blazingly fast rate.
Using a combination of genomic analysis techniques used to study the three-dimensional organization of genomes and chromatin interactions developed by Dekker and colleagues called Hi-C, high-resolution imaging, genomics, proteomics and polymer modeling, Dekker and colleagues have begun to answer these important questions.
Johan Gibcus, PhD, assistant professor of systems biology and co-first author on the study explained, “We found that these machines extrude loops and as they do so they race along the chromosomes at extremely high speeds of 2-3 kilobases per second. More importantly, we discovered a simple set of priorities that define how encounters between these fast-moving machines are resolved by the cell.”
According to Dr. Gibcus, during the mitotic phase of cell division, four different loop-extruding machines are acting simultaneously along the chromosomes: two types of cohesin machines and two types of condensin machines. During interphase, the first cohesin machine extrudes loops all along the genome. As cells enter mitosis, however, a condensin machine also begins extruding loops. When a condensin machine inevitably runs into a cohesin machine, the condensin machine removes the cohesin machine from the chromosome and continues merrily on its way, extruding loops along the genome.
Next, a second type of cohesin machine holds the two sister chromatids together, about once every million bases along the genome. When a condensin machine encounters these second cohesin machines, it simply steps over it and keeps extruding. As a result, sister chromatids remain connected to each other at the apex of the loops created by the condensin machines.
Finally, when condensin machines run into any other condensin machines while extruding loops, they simply stop and hold. As a result, each chromatid is folded as a consecutive array of loops.


