About the Department of Systems Biology
One of the defining features of living organisms is their astonishing complexity. Even seemingly simple single cell organisms such as microbes display exceedingly complex behaviors, determined by intricate molecular networks in which large numbers of molecular components, pathways and chemical reactions act together. These behaviors have fascinated scientists for decades and include development, response to pathogenic and environmental insults and interactions with other organisms. Understanding how complexity of living systems arises and coordinates cellular function and pathologies continues to be one of the principal goals of biomedical research today. Read more about how the Department of Systems Biology tackles these questions on our Research and About pages.
The Department of Systems Biology (DSB) studies how biological complexity can be derived and understood from the interplay between individual components and processes that make up living organisms.
For information about our Graduate and Summer Undergraduate Programs as well as the application process, please see our Education Page.
DSB Spotlight

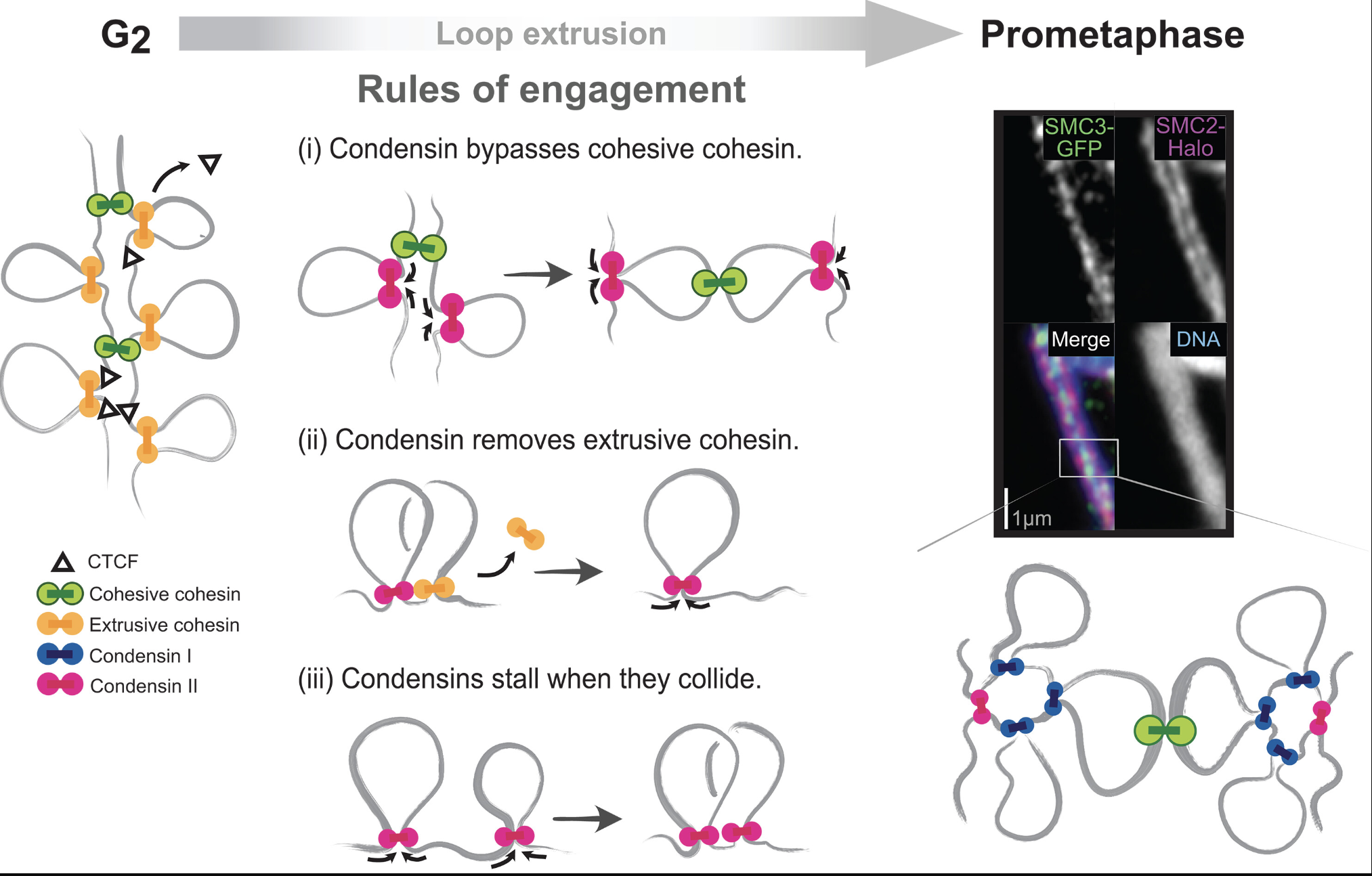
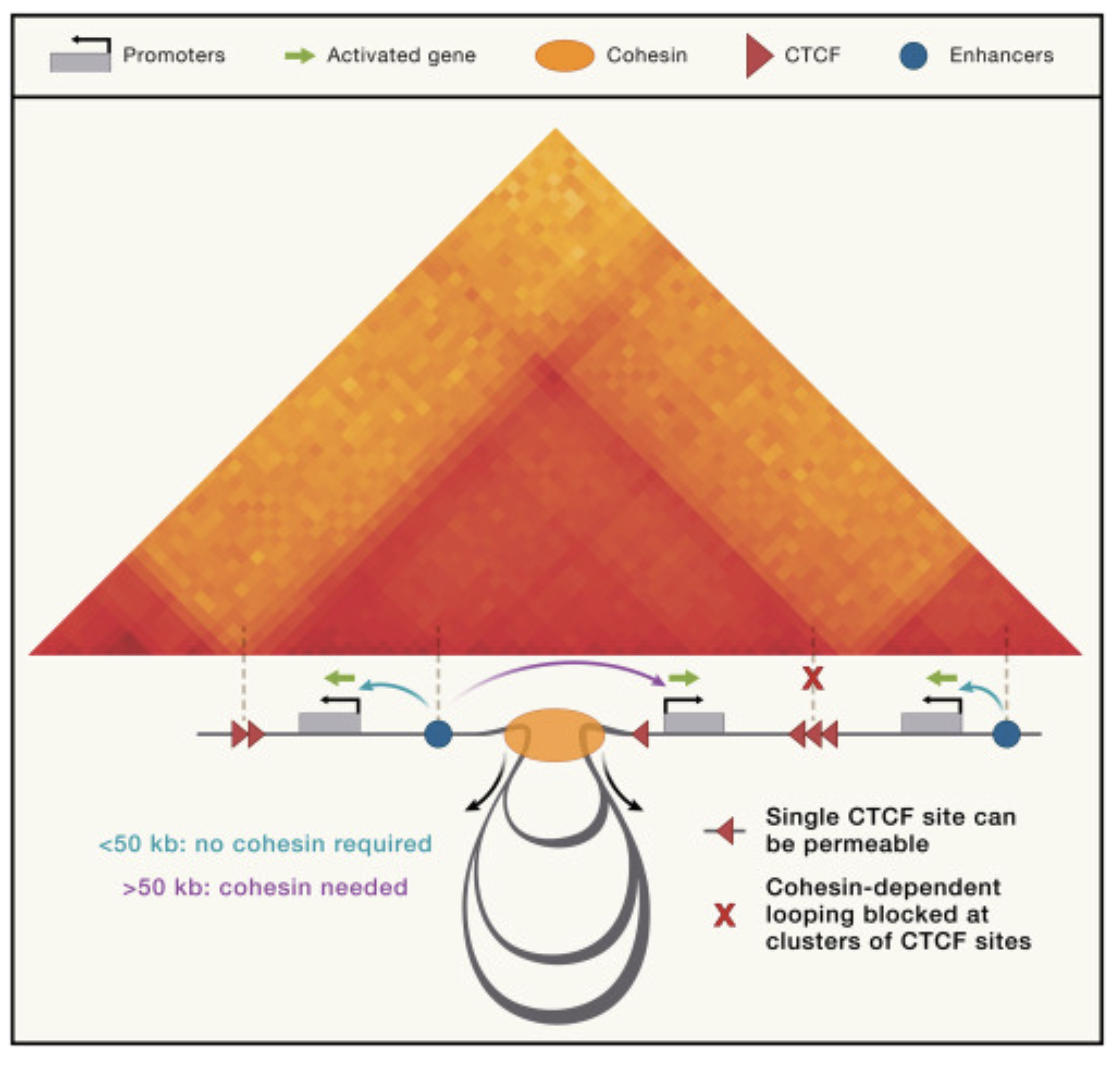
In their recent Science paper, Johan Gibcus, Job Dekker and colleagues identify rules that govern the interplay between two motors necessary for shaping chromosome structure during cell division. Read more about this work, exciting moments in the project, and Johan here.
Read the paper:
Rules of engagement for condensins and cohesins guide mitotic chromosome formation
Follow the rules in this summary video: Rules of Engagement
Article from UMASS Chan News: UMass Chan scientist co-leads study identifying simple rules for folding the genome
Upcoming Seminars
| Bart Deplancke, EPFL |
| Tuesday, May 13, 2025, 1pm (AS6-2072) |
| Host: Marian Walhout |
|
Recent Publications |
|
Rules of engagement for condensins and cohesins guide mitotic chromosome formation |
| Follow the rules in this summary video: Rules of Engagement |
| Science. 2025 April 11 |
| Kumiko Samejima, Johan H. Gibcus, Itaru Samejima, Alison J. Beckett, Nina Puǎčeková, Maria Alba Abad, Christos Spanos, Bethan Medina-Pritchard, James R. Paulson, Linfeng Xie, A. Arockia Jeyaprakash, Ian A. Prior, Leonid A. Mirny, Job Dekker, Anton Goloborodko, William C. Earnshaw |
|
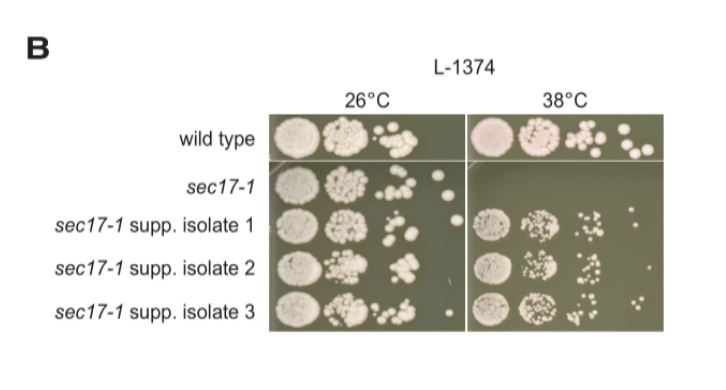
Genetic suppression interactions are highly conserved across genetically diverse yeast isolates |
| G3. 2025 March 3 |
| Claire Paltenghi, Jolanda van Leeuwen |
|
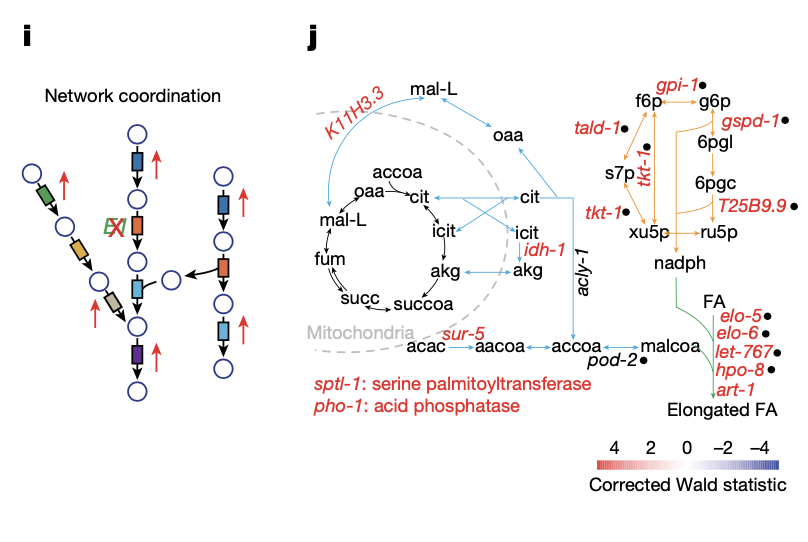
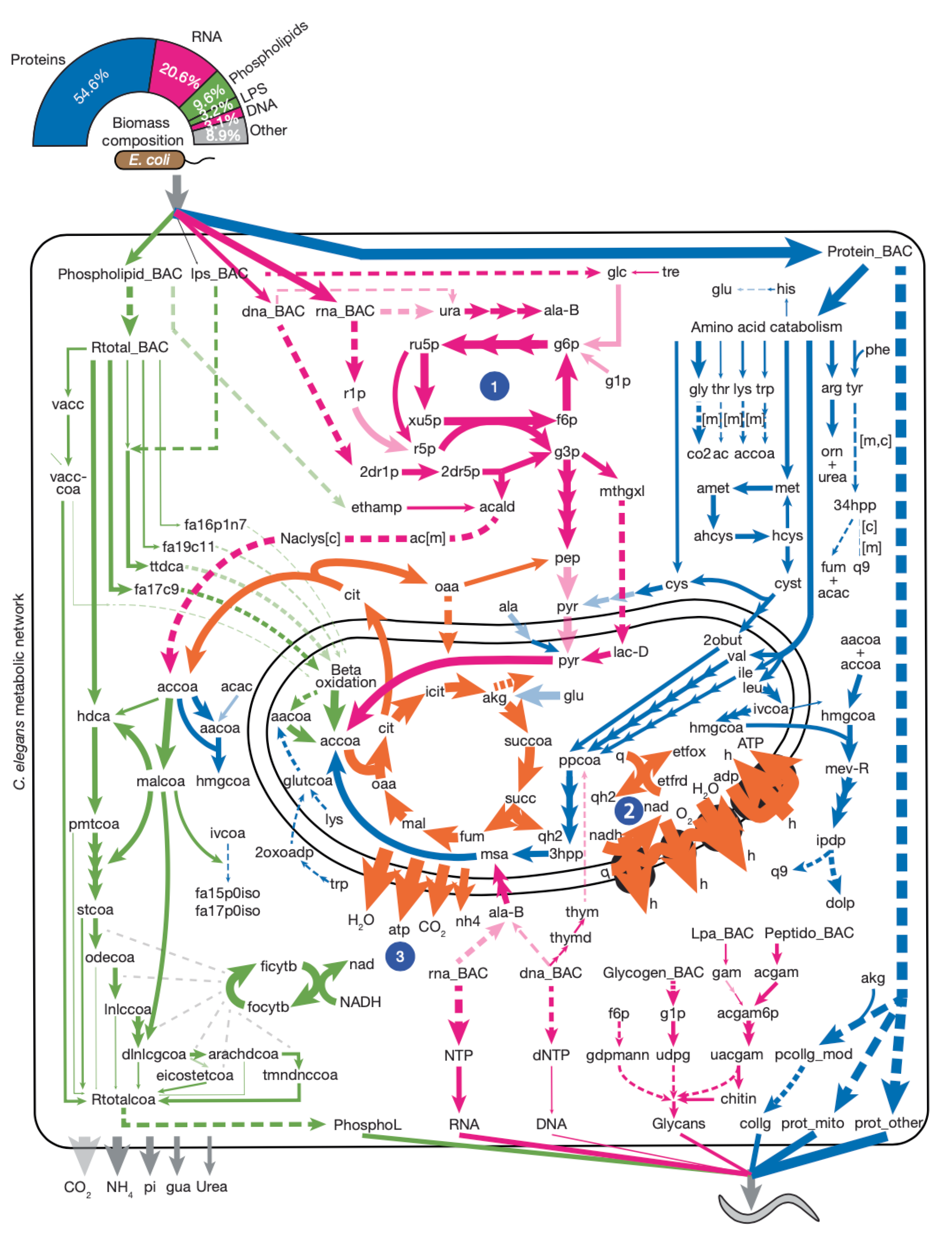
Systems-level design principles of metabolic rewiring in an animal |
| Nature. 2025 February 26 |
| Xuhang Li, Hefei Zhang, Thomas Hodder, Wen Wang, Chad L. Myers, L. Safak Yilmaz, Albertha J.M. Walhout |
|
A systems-level, semiquantitative landscape of metabolic flux in C. elegans |
| Nature. 2025 February 26 |
| Hefei Zhang, Xuhang Li, L. Tenzin Tseyang, Gabrielle E. Giese, Hui Wang, Bo Yao, Jingyan Zhang, Rachel L. Neve, Elizabeth A. Shank, Jessica B. Spinelli, L. Safak Yilmaz, Albertha J.M. Walhout |
|
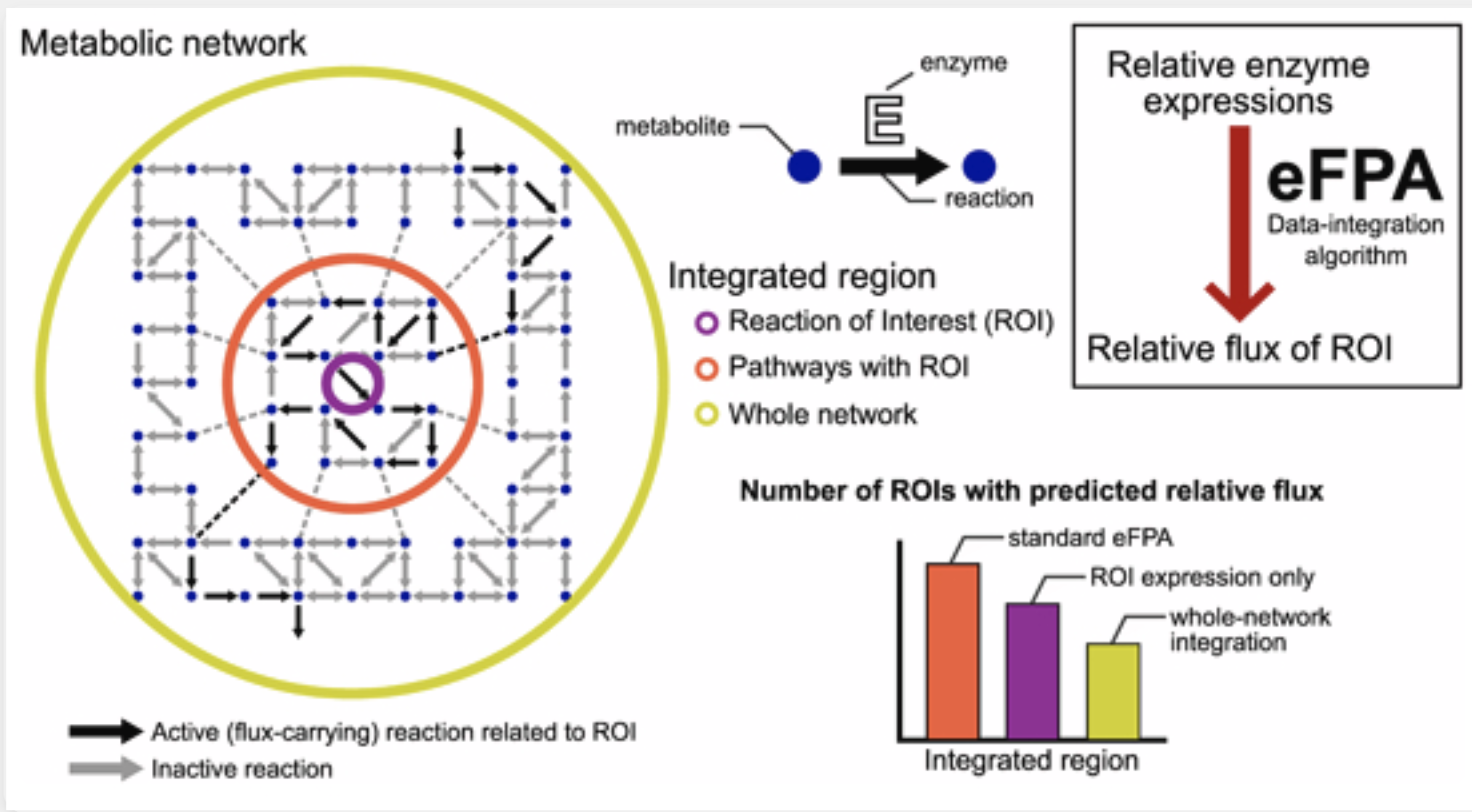
Enhanced flux potential analysis links changes in enzyme expression to metabolic flux |
| Molecular Systems Biology. 2025 February 17 |
| Xuhang Li, Albertha J.M. Walhout, L. Safak Yilmaz |
|
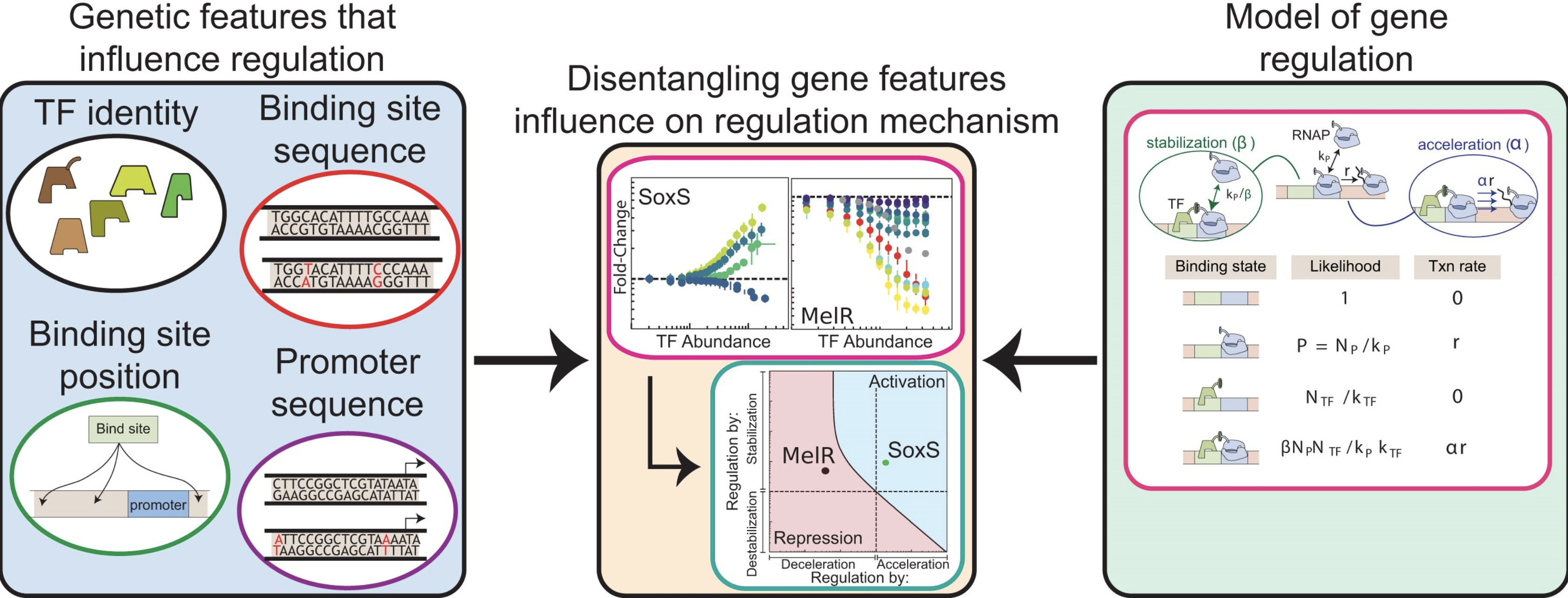
|
| Nucleic Acids Research. 2025 February 8 |
| Sunil Guharajan, Vinuselvi Parisutham, Robert C Brewster |
|
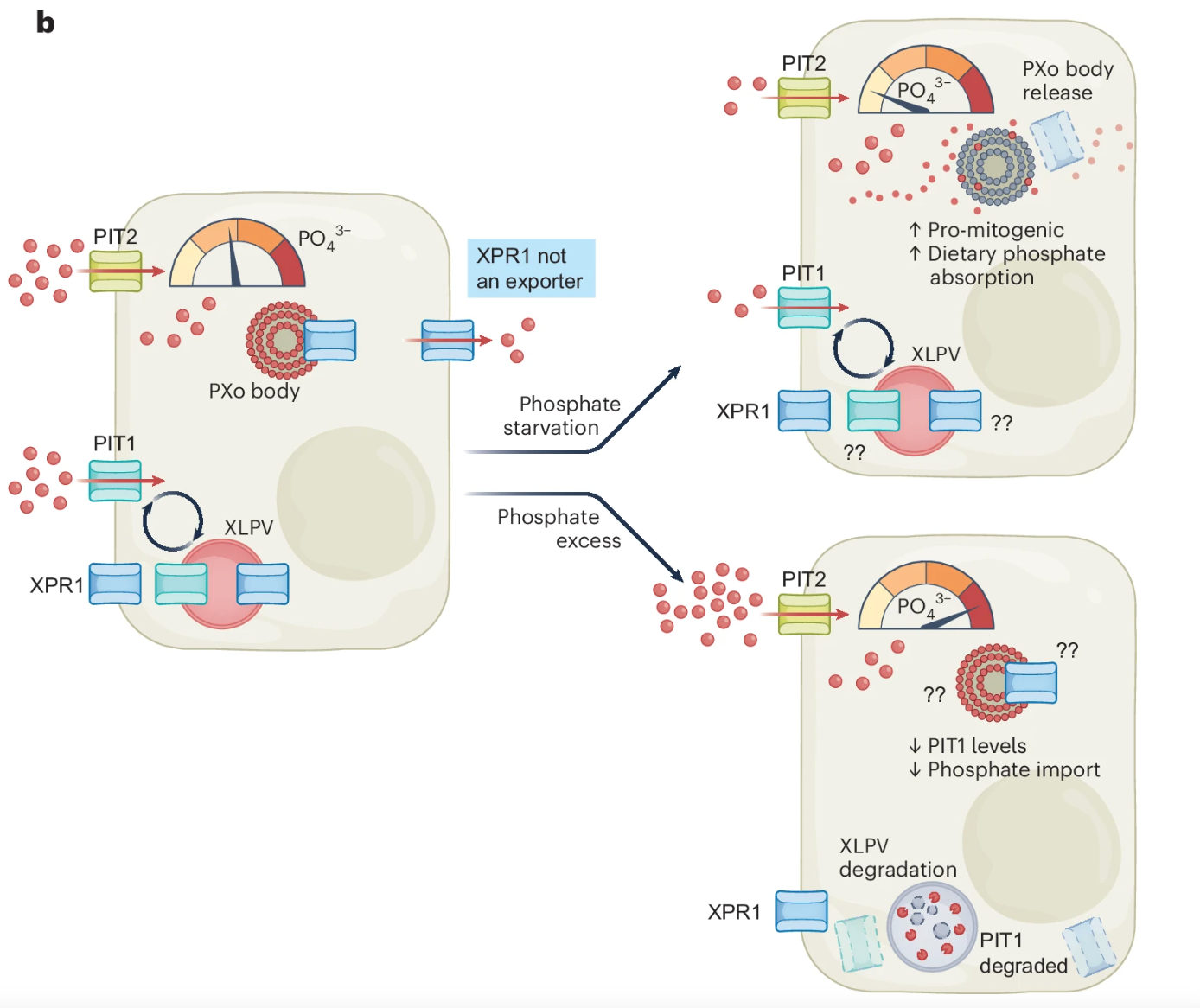
|
| Nature Structural & Molecular Biology. 2024 December 30 |
| Daniel P. Bondeson |
|
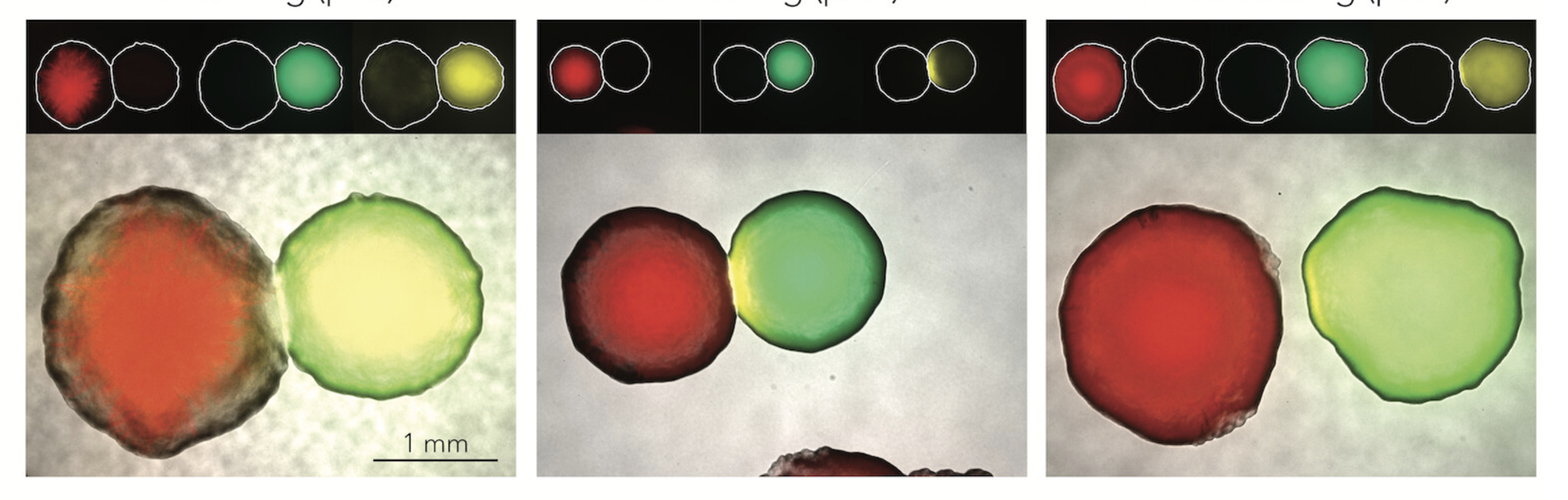
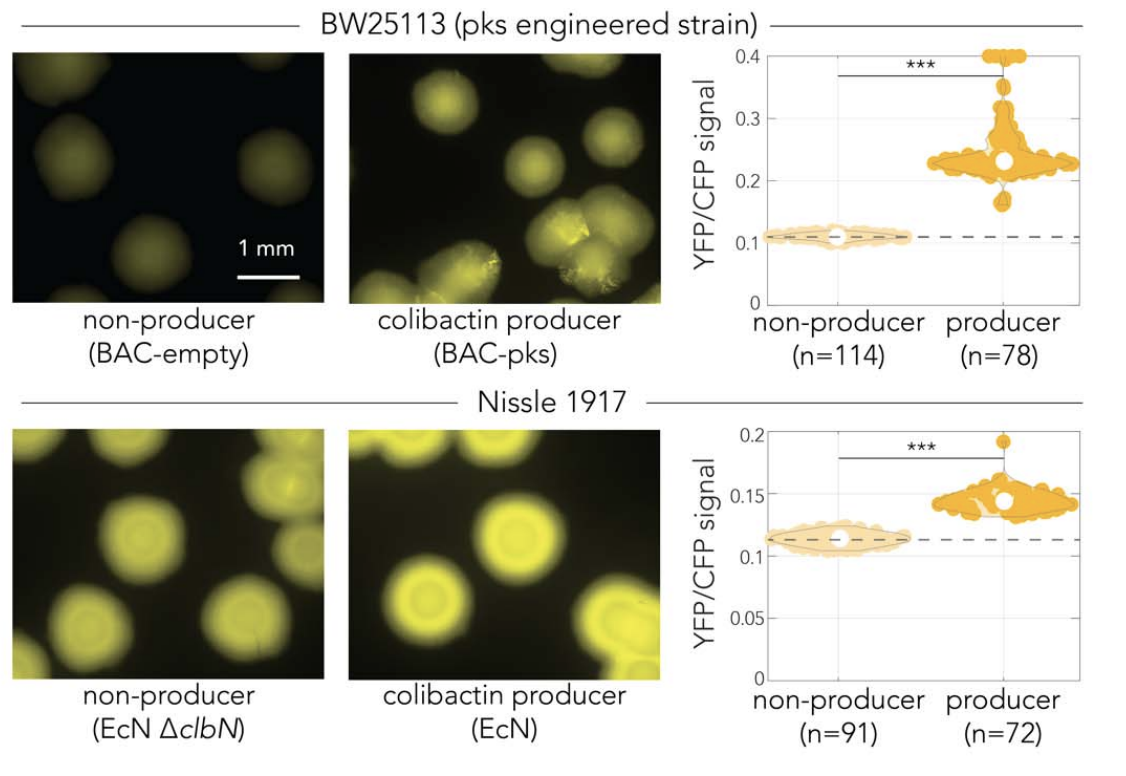
Colibactin-induced damage in bacteria is cell contact independent |
| mBio. 2024 November 22 |
| Emily Lowry and Amir Mitchell |
|
|
| Cell. 2024 November 14 |
| Job Dekker and Leonid A. Mirny |
|
Colibactin leads to a bacteria-specific mutation pattern and self-inflicted DNA damage |
| Genome Research. 2024 August 16 |
| Emily Lowry, Yiging Wang, Tal Dagan, Amir Mitchell |
| |
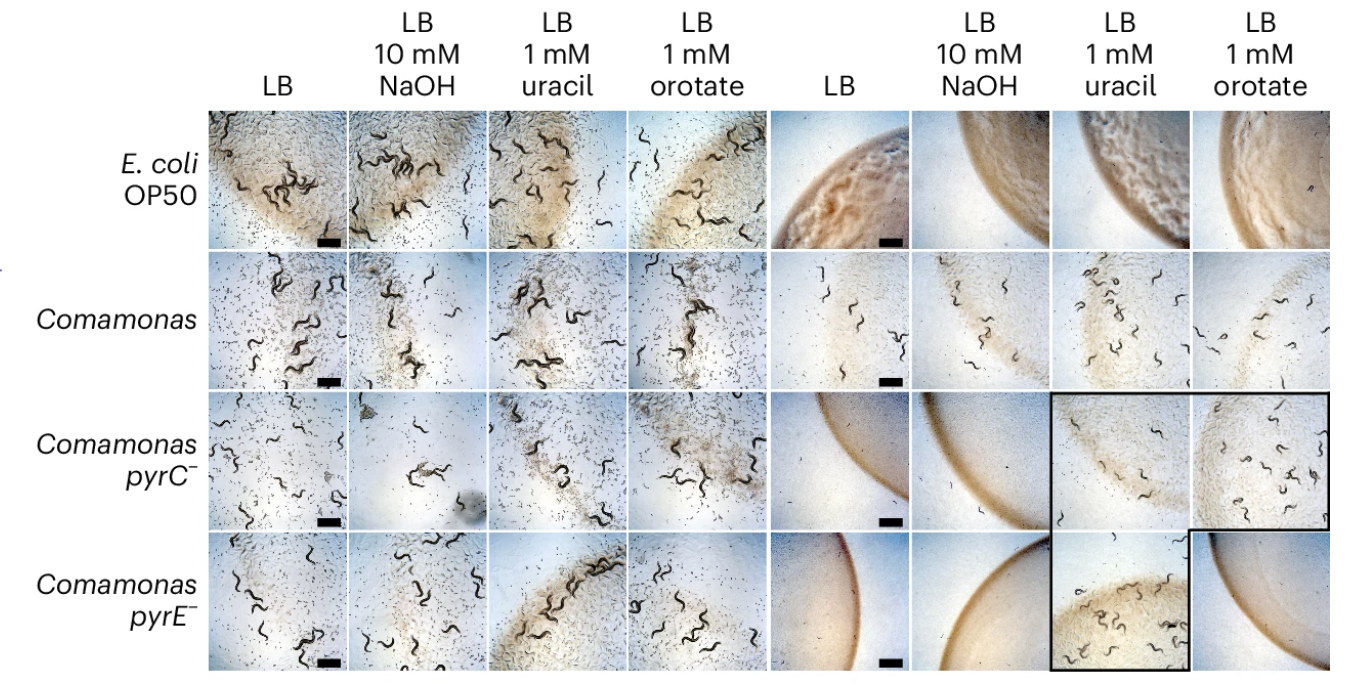
Host-microbe interactions rewire metabolism in a C. elegans model of leucine breakdown deficiency |
| Nature Metabolism. 2024 August 8 |
| Yong-Uk Lee, Bennett W. Fox, Rui Guo, Brian J. Curtis, Jingfang Yu, Sookyung Kim, Shivani Nanda, Victor Baumann, L. Safak Yilmaz, Cole M. Haynes, Frank C. Schroeder, Albertha J.M. Walhout |
| |